Pleiotropy
Below, we present the genetic correlations between phenotypes that we predict from DNA.
Genetic correlation (rg) measures how much the genetic factors influencing one trait overlap with those influencing another trait. It ranges from -1 to 1, where:
- rg = 1: The same genetic variants fully explain both traits.
- rg = 0: No shared genetic basis between the traits.
- rg = -1: Genetic variants have opposite effects on the two traits.
We use LD Score Regression (LDSC) to estimate rg using summary statistics from genome-wide association studies (GWAS).
For now, we are only using GWAS of UK Biobank participants, but we plan to include GWAS from other large-scale studies in the near future.
In simple terms, LDSC-derived rg tells us whether the same genetic influences drive two traits (e.g., height and weight), potentially
indicating a shared biological mechanisms.
The genetic correlation indicates the degree to which genetic factors influencing one trait overlap with those affecting another.
However, interpreting rg requires caution due to the following considerations:
Pleiotropy as a Cause: Genetic correlation may arise due to pleiotropy, where a single genetic variant influences multiple traits.
For example, some of the changes to DNA that causes triglyceride levels to be higher may also contribute to lower HDL cholesterol,
leading to a negative rg between these two traits.
This reflects a shared biological mechanism.
Environmental Influences: The observed rg can be conditional on the environment.
Environmental factors (e.g., societal practices practices) can modulate how genetic variants manifest,
potentially inflating or suppressing rg.
For instance, a genetic correlation between year of first live birth (in women) and fluid intelligence score
might appear in one society, in which women who do better in school are more likely to delay family formation to pursue higher education.
In another society in which all women begin family formation at the same time, that genetic correlation might vanish.
Select two traits to view their genetic correlation
Clustering traits by genetic correlation
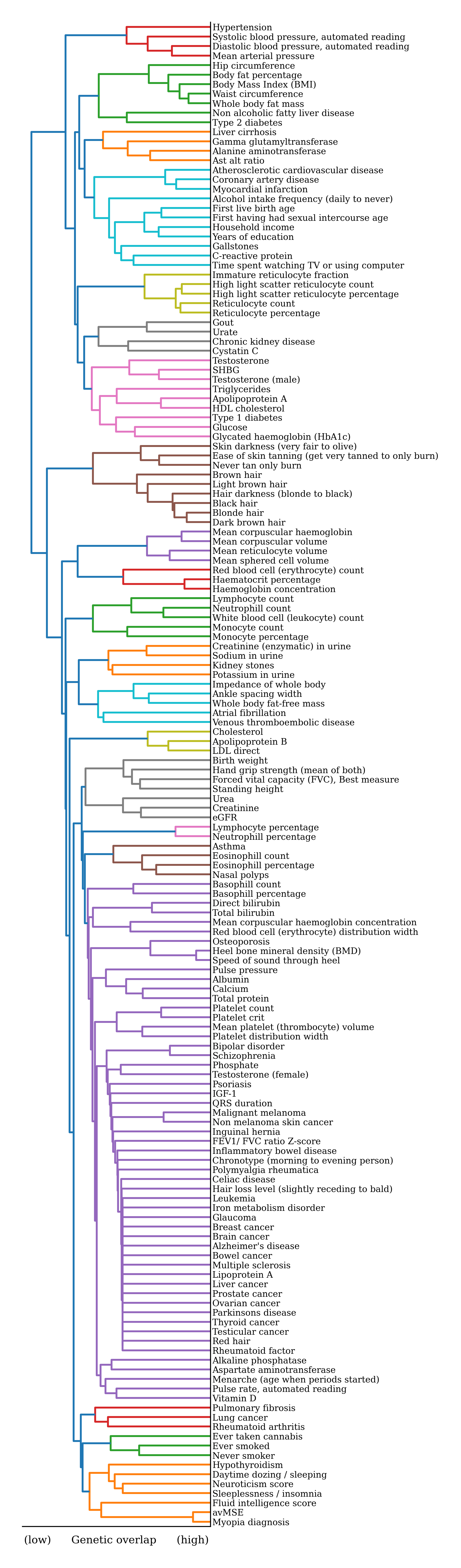
Below is a hierarchical clustering of the genetic correlations of the 158 traits that we predict from DNA. We set the genetic correlation to zero between two traits that do not reach study-wide statistical significance. That way, we can be confident that a cluster pinpoints traits that likely share similar genetic etiology and reflects underlying pleiotropy.There is no perfect clustering algorithm, the method deployed here favors conservative estimates of groups of traits that have strong genetic overlap, but may mask strong correlations between traits in different groups. Many of the traits in the purple cluster have no clear genetic overlap with each other and are lumped together as a set of singleton traits. It is worth clarifying that in the chart below, the order of the traits on the right-hand side is meaningless, it's the distance to their branching point that represents the similarity between a pair of traits.
Despite its weaknesses, this approach reveals some clusters that share interconnected genetic architectures. For example, we see a skin and hair tone cluster, a bone health cluster (osteoporosis and bone density), and an interesting genetic overlap of fluid intelligence score and myopia. The trait in that cluster, avMSE, is the average mean spherical equivalent, which measures how the eye's optical system deviates from perfect focus. A negative MSE indicates myopia (nearsightedness), where distant objects are blurry, while a positive MSE indicates hyperopia (farsightedness), where near objects may be blurry. A value close to zero suggests normal vision.
An identified cluster of diseases and biomarkers can be leveraged by constructing a multitrait polygenic predictor that borrows statistical power across multiple disease outcomes and biomarkers that share an underlying genetic architecture, e.g. heart attack, hypertension, and HDL cholesterol. In this approach, instead of an individual being provided a percentile for a specific disease outcome, they are provided a percentile for the general factor of health for some etiological category, e.g., cardiovascular, autoimmune, or kidney-related diseases. It has been shown by the Taiwan Precision Medicine Initiative that such a multitrait model achieves better predictive performance for a specific disease outcome, e.g. heart attack, than does the genetic predictor based on that disease outcome alone.